The sequencing of the genetic information of human genome has become affordable due to high-throughput sequencing technology. This opens new perspectives for the diagnosis and successful treatment of cancer and other genetic illnesses. However, there remain challenges, scientific as well as computational, that need to be addressed for this technology to find its way into everyday practice in healthcare and medicine. The first challenge is to cope with the flood of sequencing data (several Terabytes of data per week are currently collected by Vital-IT (the SIB's High Performance and High Throughput Bioinformatics Competency Center). For instance, a database covering the inhabitants of a small country like Switzerland would need to store a staggering amount of data, about 2'335'740 Terabytes. The second challenge is the ability to process such enormous deluge of data in order to 1) increase the scientific knowledge of genome sequence information and 2) search genome databases for diagnosis and therapy purposes. Significant compression of genomic data is required to reduce the storage size, to increase the transmission speed and to reduce the cost of I/O bandwidth connecting the database and the processing facilities. In order to process the data in a timely and effective manner, algorithms need to exploit the significant data parallelism and they need to be able to "scale" and "map" the data processing stage on the massive parallel processing infrastructure according to the available resources and to the application constraints.
The main aim of the project is to develop for the Swiss Platform for Advanced Scientific Computing a new computation node composed of heterogeneous hardware, a new compression format for genomic data and a software infrastructure that enables emerging applications such as Genome Analysis (e.g. RNASeq) to be able to process extremely large volumes of genome data in an efficient and timely way for scientific and diagnostic purposes (clinical application) as depicted in Figure 1. Specifically, we will select a representative software application in the domain of RNASeq (e.g. tophat, bowtie or similar), and provide a high performance hardware/software implementation. It will then serve as an example for other applications.
The project will achieve the goal by addressing the following issues and tasks:
- Select existing genomics applications such as tophat and bowtie as essential components for a typical genome analysis application.
- Data compression and decompression:
- select existing compression algorithms that, beside compression, provide the appropriate functionality for the efficient implementation of analysis applications such as tophat and bowtie in terms of reduced analysis time
- basing on the assessment of the performance of existing compression algorithms and their efficient implementation, design new improved algorithms
- Computational engines:
- the different software components necessary to implement the genome analysis programs accessing compressed genomes will be implemented using a portable high level dataflow programming methodology
- a set of tools that synthesize and profile the high level dataflow programs to build genome analysis applications that efficiently runs on the PASC processing infrastructure will be developed
- platform specific libraries with the potential of supporting further algorithmic implementations will be developed.
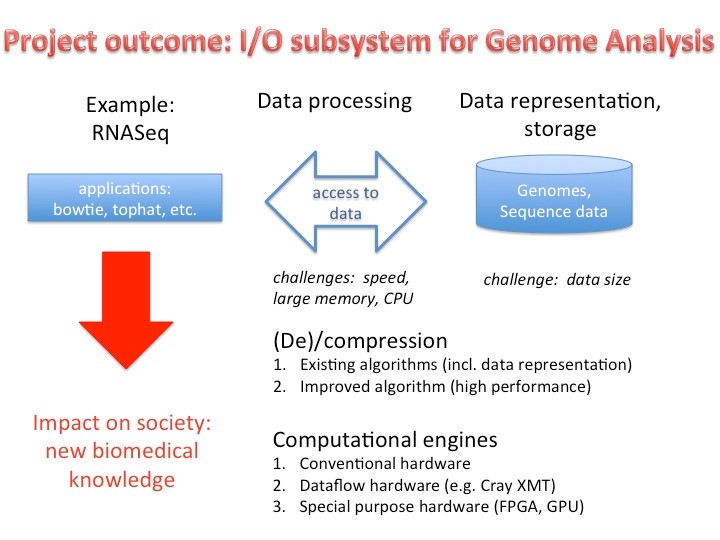
Figure 1. Project outcome and application area.
This project will address the various challenges with the following technical thrusts:
- Investigate the realization of very high-throughput I/O solutions embedding genome compression/decompression algorithm implementations as hardware co-processing units. Compression being based on entropy coding, its essentially serial and bitwise structure is a poor match for current processor instruction sets and therefore requires specific optimized implementations. A platform architecture will be proposed and discussed with the CSCS team in order to fit their future computation node.
- Re-factor genome analysis and processing algorithms and associated applications to benefit from the portable parallelism paradigm resulting from a dataflow programming approach. This work is fundamental not only for obtaining acceptable levels of application performance, but also for exploiting the evolution of processing capabilities. Changes in the processing infrastructure necessitate porting processing algorithms, and the portability of dataflow programs promises to overcome the need to re-write applications when higher or different forms of parallelism and concurrency will be available. This task will also include embedding of processing-intensive functions into the (high level) genome application software which, due to their “bitwise” nature, require processor “native” implementations or hardware co-processing. Finally, the new algorithms will have to scale and adapt to the future CSCS node, notably in terms of memory size, number of nodes, and types of nodes.
- Further develop the dataflow programming methodology and associated ecosystem of tools built around the dataflow programming standard ISO/IEC 23001-4 so far used essentially in the video streaming and compression field so that it can become the development environment of genome application software and other domain applications that can benefit its features such as portable parallelism.
Overall, this proposal is in the domain of data-intensive memory-bound applications that further pushes the requirements of special purpose hardware. Our applications seem to be a perfect match for non-standard hardware architectures and the development of high-performance algorithms and libraries.
Within that context the REDS institutes is involved in the acceleration of both compression/decompression and sequences comparison. FPGAs will be exploited in order to speed up these processings.
Nowadays, in order to analyze and read the human genome, sequencers must split the DNA chain into small fragments called sequences. There is a huge redundancy in the sequences produced by a DNA sequencer machine, implying an high coverage of the analyzed genome. One method to compress the output data of a sequencer is therefore to take advantage of this data redundancy.
One main step into the genomic data compression process is the detection of identical (or more or less identical) sequences. This task, called clustering, aims to group together every similar sequences (or with a small offset). The main challenge during the clustering phase is to speed up as much as possible the match between two sequences, since this operation has to be performed on several millions of DNA fragments each of which is composed of a few hundreds bases.
A base can be represented by a letter : A,C,G,T or N (where N is unknown). Three bits are therefore enough to encode each possible type of bases within a sequence. It has to be noted that this fine granularity fits perfectly with the FPGAs capability.

The aim of the current work is to develop a system capable of drastically speed up the clustering process presented in the previous paragraphs. In order to achieve this goal the design will be implemented on a FPGA based hardware connected to a standard PC via a PCIe link.
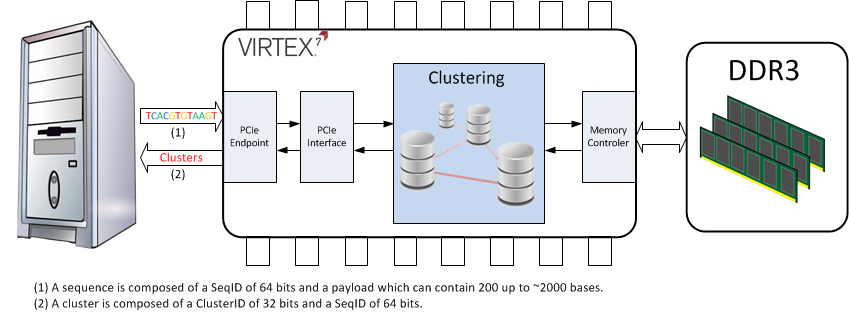
The main features of this subproject are :
The system shall be able to handle sequences with a variable length The memory presented on the FPGA board shall be used in order to cache the data The system will be designed to handle a new type of memory elements called HMC (Hybrid Memory Cube)
At the end, the performances of this system shall be compared with those provided by a standard clustering software.